Say a graph’s vertex set is and edge set is
. We want to traverse the graph efficiently, therefore, we want to visit each vertex and each edge exactly once.
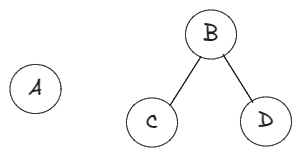
An edge has two ends: a from and a to. In the edge we can consider
as
from and as
to. The basic traversal strategy is to consider each vertex in the from role exactly once and look at all its to‘s to find not-yet-considered from roles. To avoid putting a vertex in the from role twice, as soon as we decide to consider it in the from role, we mark it as visited.
So, during traversal is partitioned into two subsets: visited and not-visited:

We can visit the “not-visited” subset in many different orders. Two among these orders enable us to efficiently answer different questions about the graph: DFS and BFS.
DFS
We visit not-visited vertices as we find them and backtrack only when there are no more not-visited to‘s.
DFS can explore a graph in time. Piggybacking on the
precompute() or postcompute() in DFS, we can solve various problems.
def explore(u, adjacency_list, visited) -> None:
visited[u] = True
precompute()
for v in adjacency_list[u]:
if visited[v]:
continue
explore(v, adjacency_list, visited)
postcompute()
def dfs(adjacency_list: List[List[int]]) -> None:
n = len(adjacency_list)
visited = [False] * n
for u in range(n):
if visited[u]:
continue
explore(v, adjacency_list, visited)
Is undirected graph connected?
Check if all vertices belong to a single connected component.
Time: . Space:
.
def dfs(u: int, adj_list: List[List[int]], visited: List[bool], ccnum: List[int], cc: int) -> None:
visited[u] = True
ccnum[u] = cc
for v in adj_list[u]:
if visited[v]:
continue
dfs(v, adj_list, visited, ccnum, cc)
def find_connected_components(adj_list: List[List[int]]) -> List[int]:
n = len(adj_list)
ccnum, cc = [0] * n, 0
visited = [False] * n
for u in range(n):
if visited[u]:
continue
dfs(u, adj_list, visited, ccnum, cc)
cc += 1
return ccnum
def is_connected_undirected(adj_list: List[List[int]]) -> bool:
ccnum = find_connected_components(adj_list)
return len(set(ccnum)) == 1
adj_list = [
[1, 4], # 0
[0], # 1
[3, 6, 7], # 2
[2, 7], # 3
[8, 9], # 4
[], # 5
[2, 7, 10], # 6
[2, 3, 10, 11], # 7
[4, 9], # 8
[4, 8], # 9
[6, 7], # 10
[7] # 11
]
connected = is_connected_undirected( adj_list )
print(f"{connected=}")
adj_list2 = [
[1, 2], # 0
[0], # 1
[0, 3], # 2
[0, 2] # 3
]
connected = is_connected_undirected( adj_list2 )
print(f"{connected=}")
# connected=False
# connected=True
Categorize directed edges
A directed edge can belong to one of four categories based on when
sees
during DFS.
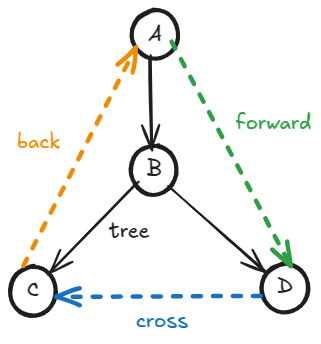
- Tree edge:
is a child of
. The vertex
had not been visited when
saw it. If the edge
represents that the task
depends on the task
, then
must be done before we can do
. The vertex
is about to be put in the recursion call stack.
- Forward edge:
is a descendant of
. The vertex
had already been entered and exited — therefore completely processed. The vertex
is not in the recursion call stack.
- Back edge:
is an ancestor of
. The vertex
had been entered but not exited yet. Therefore,
is still in the call stack.
- Cross edge:
is neither descendant nor ancestor of
. The vertex
has been entered and exited — therefore completely processed;
is not in the call stack. Additionally, down from their least common ancestor, the subtrees
and
belong to, could have been processed concurrently without breaking dependency constraints.
From DFS pre- and post-times, we can categorize the edges.
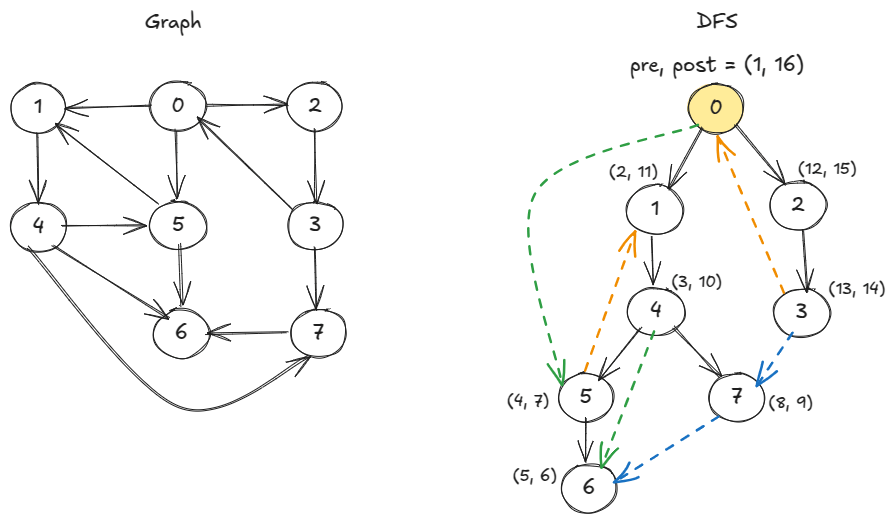
def dfs(u, adj_list, visited, clock, pre, post) -> None:
visited[u] = True
clock[0] += 1
pre[u] = clock[0]
for v in adj_list[u]:
if visited[v]:
continue
dfs(v, adj_list, visited, clock, pre, post)
clock[0] += 1
post[u] = clock[0]
def compute_pre_post_time(adj_list: List[List[int]]) -> Tuple[List[int], List[int]]:
n = len(adj_list)
visited = [False] * n
pre, post = [0] * n, [0] * n
clock = [0]
for u in range(n):
if visited[u]:
continue
dfs(u, adj_list, visited, clock, pre, post)
return pre, post
def categorize_edges(adj_list: List[List[int]]) -> Dict[str, List[List[int]]]:
pre, post = compute_pre_post_time( adj_list )
n = len(adj_list)
cat = defaultdict(list)
for u in range(n):
for v in adj_list[u]:
edge = [u, v]
if pre[u] < pre[v] < post[v] < post[u]:
cat["tree_or_forward"].append( edge )
elif pre[v] < pre[u] < post[u] < post[v]:
cat["back"].append( edge )
else:
cat["cross"].append( edge )
return cat
adj_list = [
[1, 2, 5], # 0
[4], # 1
[3],
[0, 7],
[5, 6, 7],
[1, 6],
[],
[6]
]
cat = categorize_edges( adj_list )
print(f"{cat=}")
# cat=defaultdict(<class 'list'>, {'tree_or_forward': [[0, 1], [0, 2], [0, 5], [1, 4], [2, 3], [4, 5], [4, 6], [4, 7], [5, 6]], 'back': [[3, 0], [5, 1]], 'cross': [[3, 7], [7, 6]]})
Is directed graph acyclic?
A directed graph has a cycle if and only if DFS finds a back edge.
During DFS, for an edge , if
has already been entered but not yet exited and
, then
is a back edge. Or, if
has already been visited and it is still in the stack (
), it is a back edge.
# def is_back_edge(u, v, pre, post) -> bool:
# # v has not been entered yet
# if pre[v] == 0:
# return False
# # v has already been exited
# if post[v] != 0:
# return False
# # v is not an ancestor
# if pre[v] > pre[u]:
# return False
# return True
def is_back_edge(u: int, v: int, visited: List[bool], post: List[int]) -> bool:
return visited[v] and post[v] == 0
def has_back_edge_dfs(u, adj_list, visited, clock, pre, post) -> bool:
visited[u] = True
clock[0] += 1
pre[u] = clock[0]
for v in adj_list[u]:
if is_back_edge(u, v, pre, post):
return True
if visited[v]:
continue
if has_back_edge_dfs(v, adj_list, visited, clock, pre, post):
return True
clock[0] += 1
post[u] = clock[0]
return False
def has_directed_cycle(adj_list: List[List[int]]) -> bool:
n = len(adj_list)
visited = [False] * n
clock = [0]
pre, post = [0]*n, [0]*n
for u in range(n):
if visited[u]:
continue
if has_back_edge_dfs(u, adj_list, visited, clock, pre, post):
return True
return False
adj_list_with_cycle = [
[1, 2, 5], # 0
[4], # 1
[3],
[0, 7],
[5, 6, 7],
[1, 6],
[],
[6]
]
has_cycle = has_directed_cycle( adj_list_with_cycle )
print(f"{has_cycle=}")
adj_list_without_cycle = [
[2],
[0, 3],
[4, 5],
[2],
[],
[]
]
has_cycle = has_directed_cycle( adj_list_without_cycle )
print(f"{has_cycle=}")
# has_cycle=True
# has_cycle=False
Topological sort
By DFS post
For a DAG, vertices in decreasing order of DFS post is a topologically sorted (or linearized) order. If we process the vertices in this order, edges will always flow from earlier to later vertices.
def explore(u: int, adj_list: List[List[int]], visited: List[bool], topo_order: List[int]) -> None:
visited[u] = True
for v in adj_list[u]:
if visited[v]:
continue
explore(v, adj_list, visited, topo_order)
topo_order.append(u)
def dfs(adj_list, topo_order: List[int]) -> None:
n = len(adj_list)
visited = [False] * n
for u in range(n):
if visited[u]:
continue
explore(u, adj_list, visited, topo_order)
def topo_sort(adj_list: List[List[int]]) -> List[int]:
topo_order = []
dfs(adj_list, topo_order)
# reverse to have FIFO: decreasing order of post
return topo_order[::-1]
By in-degree
For a directed edge , we can think
as a pre-condition on
. Then the vertices with in-degree = 0, do not have any pre-condition and we can process them immediately. Lets call them sources. Once we process a source, we can delete it and all its edges. That causes in-degrees of other vertices to drop and some of these become new sources. Processing vertices in increasing order of when they became “sources,” gives a topological order.

Time: , extra space for in-degree tracking is
.
def topo_sort_by_in_degree(adj_list: List[List[int]]) -> List[int]:
n = len(adj_list)
# Sources will not appear in any edge-list, so init
# in-degree with 0.
in_degree = defaultdict(int, { u: 0 for u in range(n) })
for u in range(n):
for v in adj_list[u]:
in_degree[v] += 1
# Vertices without preconditions
sources = [u for u, d in in_degree.items() if d == 0]
topo_order = []
while sources:
u = sources.pop()
topo_order.append(u)
for v in adj_list[u]:
in_degree[v] -= 1
if in_degree[v] == 0:
sources.append(v)
return topo_order
On each iteration, we can process all sources concurrently. It is like peeling an onion.
def topo_sort_indegree_levelwise(adj_list: List[List[int]]) -> List[int]:
n = len(adj_list)
indegree = defaultdict(int, {u: 0 for u in range(n)})
for u in range(n):
for v in adj_list[u]:
indegree[v] += 1
curr_src = [u for u, d in indegree.items() if d == 0]
topo_order = []
while curr_src:
topo_order.extend(curr_src)
new_src = []
for u in curr_src:
for v in adj_list[u]:
indegree[v] -= 1
if indegree[v] == 0:
new_src.append(v)
curr_src = new_src
return topo_order
All topological orderings
We can embed in-degree approach in backtracking to find all topological orderings. At each level, we extend the current_order by one more source, picking every source available at that level as the extension choice. Once current_order includes all vertices, we copy it into the all_orderings collection.
def decrease_indegree(u, indegree, adj_list) -> None:
for v in adj_list.get(u, []):
indegree[v] -= 1
def increase_indegree(u, indegree, adj_list) -> None:
for v in adj_list.get(u, []):
indegree[v] += 1
def backtrack(indegree, adj_list, visited, curr_topo, all_topo) -> None:
if len(curr_topo) == len(adj_list):
all_topo.append( list(curr_topo) )
return
for u in sorted(adj_list):
if u in visited or indegree[u] > 0:
continue
curr_topo.append(u)
visited.add(u)
decrease_indegree(u, indegree, adj_list)
backtrack(indegree, adj_list, visited, curr_topo, all_topo)
increase_indegree(u, indegree, adj_list)
visited.remove(u)
curr_topo.pop()
def get_all_topological_orders(adj_list) -> List[List[str]]:
all_order = []
indegree = get_indegree_map(adj_list)
backtrack(indegree, adj_list, set(), [], all_order)
return all_order
adj_list_dag = {
"A": {"C"},
"B": {"A", "D"},
"C": {"E", "F"},
"D": {"C"},
"E": {},
"F": {}
}
all_topo = get_all_topological_orders(adj_list_dag)
print(f"{all_topo=}")
# all_topo=[['B', 'A', 'D', 'C', 'E', 'F'], ['B', 'A', 'D', 'C', 'F', 'E'], ['B', 'D', 'A', 'C', 'E', 'F'], ['B', 'D', 'A', 'C', 'F', 'E']]
All directed cycles
If we maintain DFS parents, as soon as DFS finds a back edge , we can find the associated cycle:
.
def tree_path(u, v, parent) -> List[str]:
path = deque()
current = v
while current != u:
path.appendleft(current)
current = parent[current]
path.appendleft(u)
return path
def explore_cycle(u, adj_list, visited, onstack, parent, cycles) -> None:
visited.add(u)
onstack.add(u)
for v in adj_list.get(u, []):
if v in visited:
if v in onstack:
# back edge: u -> v
# cycle: v -> ... -> u + u -> v
v_u_path = tree_path(v, u, parent)
v_u_path.append(v)
cycles.append( v_u_path )
continue
parent[v] = u
explore_cycle(v, adj_list, visited, onstack, parent, cycles)
onstack.remove(u)
def dfs(adj_list) -> List[List[str]]:
visited = set()
onstack = set()
parent = {}
cycles = []
for u in adj_list:
if u in visited:
continue
parent[u] = None
explore_cycle(u, adj_list, visited, onstack, parent, cycles)
return cycles
dir_adj_list_cycle = {
"A": ["B", "C", "F"],
"B": ["E"],
"C": ["D"],
"D": ["A", "H"],
"E": ["F", "G", "H"],
"F": ["B", "G"],
"H": ["G"]
}
cycles = dfs( dir_adj_list_cycle )
print(f"{cycles=}")
# cycles=[deque(['B', 'E', 'F', 'B']), deque(['A', 'C', 'D', 'A'])]
Strongly connected components of directed graph
In a strongly connected component, for every vertex-pair: , there is a directed path from
to
and a directed path from
to
. There are five strongly connected components in the below directed graph.
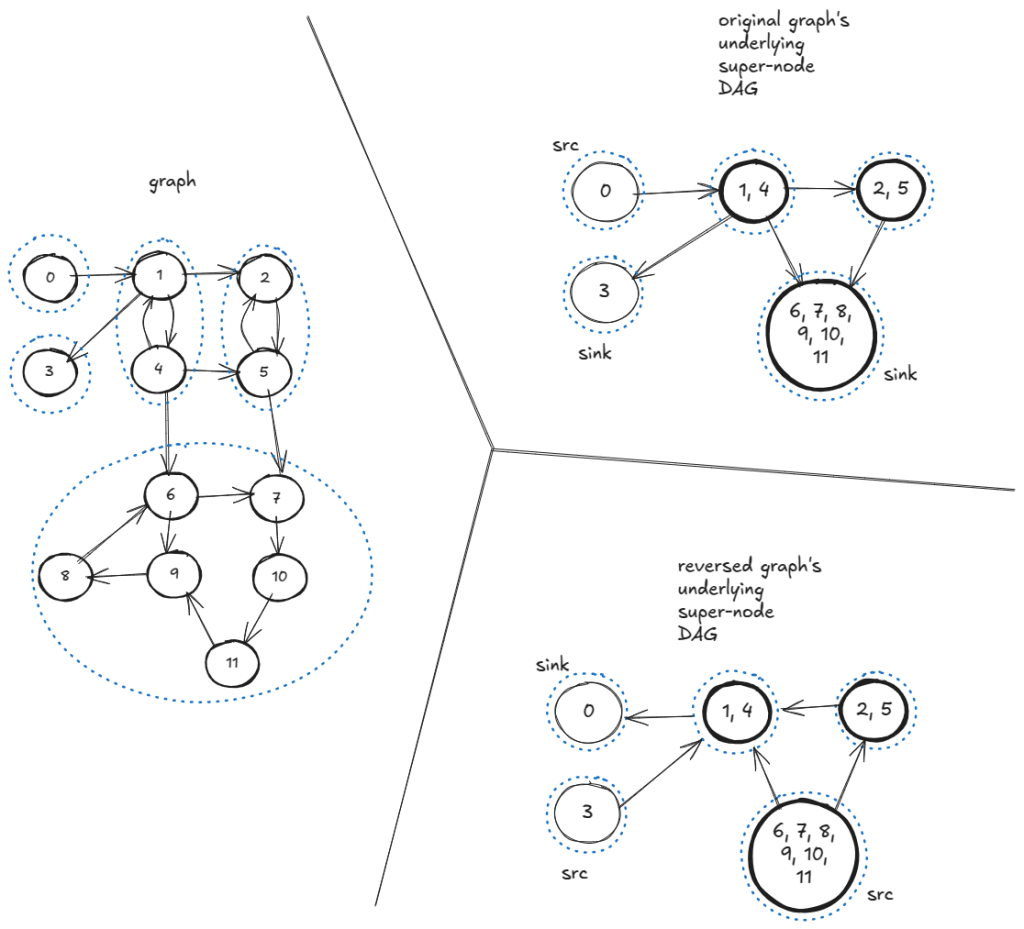
Strongly connect component . Note,
is an equivalence class for the relation
. As a result, strongly connected components partition the graph.
For every strongly connected component, if we collapse all vertices within that component into one super-node, we get a DAG of super-nodes.
Insight: In the super-node DAG, if we run DFS within a “sink” super node, the DFS will never leave that super-node or strongly connected component.
Therefore, to find strongly connected components, we can run the undirected graph connected component finding DFS here for the directed graph, but we have to process the vertices in the order of “sink” towards “src”.
The vertex having the highest DFS post time is a “src”. So, to find the “sink”, we can run DFS on the reversed graph and take the highest post in that one. In other words, running undirected connected component (cc) DFS in the decreasing post order of the reversed graph is same as running cc DFS from sink to src of the original graph.
Time and space: .
from collections import defaultdict
from typing import Dict, List
def cc_explore(u, adj_list, visited, tic, cc) -> None:
visited[u] = True
cc[u] = tic
for v in adj_list[u]:
if visited[v]:
continue
cc_explore(v, adj_list, visited, tic, cc)
def cc_dfs(adj_list, sorted_nodes) -> List[int]:
n = len(sorted_nodes)
visited = [False] * n
cc = [0] * n
tic = 1
for u in sorted_nodes:
if visited[u]:
continue
cc_explore(u, adj_list, visited, tic, cc)
tic += 1
return cc
def explore_post(u, adj_list, visited, post_order) -> None:
visited[u] = True
for v in adj_list[u]:
if visited[v]:
continue
explore_post(v, adj_list, visited, post_order)
post_order.append(u)
def decreasing_post_order(adj_list) -> List[int]:
n = len(adj_list)
visited = [False] * n
post_order = []
for u in range(n):
if visited[u]:
continue
explore_post(u, adj_list, visited, post_order)
return post_order[::-1]
def reverse_adj_list(adj_list: List[List[int]]) -> List[List[int]]:
n = len(adj_list)
rev_list = [[] for _ in range(n)]
for u in range(n):
for v in adj_list[u]:
rev_list[v].append(u)
return rev_list
def strongly_connected_components(adj_list: List[List[int]]) -> Dict[int, List[int]]:
rev_adj_list = reverse_adj_list(adj_list)
sink_to_src = decreasing_post_order(rev_adj_list)
cc = cc_dfs( adj_list, sink_to_src )
cc_map = defaultdict(list)
for u, comp in enumerate(cc):
cc_map[comp].append(u)
return cc_map
adj_list_strong_cc = [
[1], # 0
[2, 3, 4], # 1
[5], # 2
[], # 3
[1, 5, 6], # 4
[2, 7], # 5
[7, 9], # 6
[10], # 7
[6], # 8
[8], # 9
[11], # 10
[9] # 11
]
strong_cc = strongly_connected_components( adj_list_strong_cc )
print(f"{strong_cc=}")
# strong_cc=defaultdict(<class 'list'>, {5: [0], 4: [1, 4], 3: [2, 5], 2: [3], 1: [6, 7, 8, 9, 10, 11]})
BFS
We visit the not-visited nodes in a layer-by-layer manner. All adjacent to‘s first, then adjacent-adjacent to‘s etc.
def explore(src, adj_list, visited, tree):
visited.add(src)
tree.append(src)
q = deque([src])
while q:
u = q.popleft()
for v in adj_list[u]:
if v in visited:
continue
visited.add(v)
tree.append(v)
q.append(v)
def bfs(adj_list) -> List[List[str]]:
visited = set()
forest = []
for u in adj_list:
if u in visited:
continue
tree = []
explore(u, adj_list, visited, tree)
forest.append(tree)
return forest
adj_list = {
"A": ["B", "E"],
"B": ["A"],
"C": ["D", "G", "H"],
"D": ["C", "H"],
"E": ["A", "I", "J"],
"F": [],
"G": ["C", "H", "K"],
"H": ["C", "D", "G", "K", "L"],
"I": ["E", "J"],
"J": ["E", "I"],
"K": ["G", "H"],
"L": ["H"]
}
forest = bfs(adj_list)
print(f"{forest=}")
# forest=[['A', 'B', 'E', 'I', 'J'], ['C', 'D', 'G', 'H', 'K', 'L'], ['F']]
Shortest undirected, unweighted path distances from a source
Time: , space:
.
def shortest_path_distance_bfs(src, adj_list) -> Dict[str, int]:
dist = {u: float("inf") for u in adj_list}
dist[src] = 0
q = deque([src])
visited = {src}
while q:
u = q.popleft()
for v in adj_list[u]:
if v in visited:
continue
visited.add(v)
dist[v] = dist[u] + 1
q.append(v)
return dist
adj_list = {
"A": ["B", "S"],
"B": ["A", "C"],
"C": ["B", "S"],
"D": ["E", "S"],
"E": ["D", "S"],
"S": ["A", "C", "D", "E"]
}
dist = shortest_path_distance_bfs("S", adj_list)
print(f"{dist=}")
# dist={'A': 1, 'B': 2, 'C': 1, 'D': 1, 'E': 1, 'S': 0}
If we think of vertices as balls and edges as strings, BFS is like exploring the taut strings when the source-ball is pulled up.
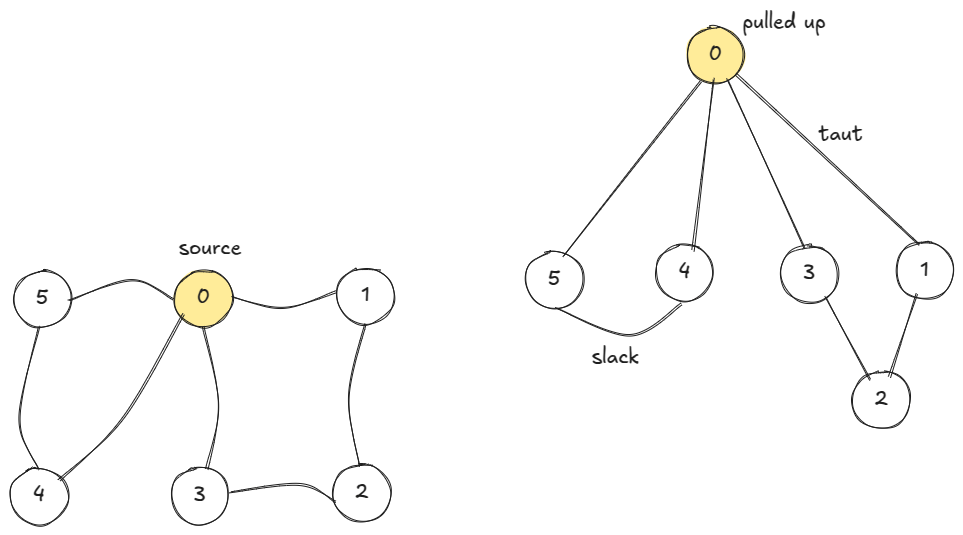
We can also think BFS as wavefront moving radially outwards from the source one unit distance in one unit time.
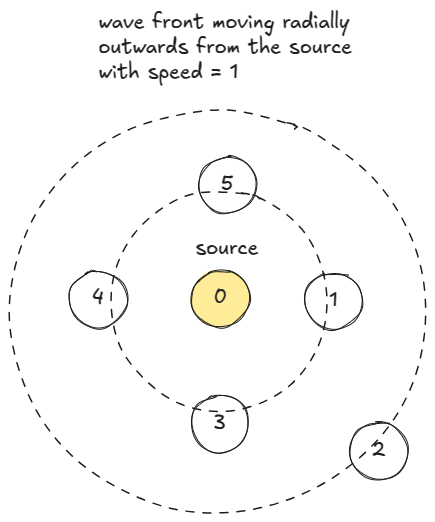
On each iteration, we can process all nodes on the wavefront concurrently.
def shortest_path_distance_bfs_wavefront(src, adj_list) -> Dict[str, int]:
dist = {u: float("inf") for u in adj_list}
dist[src] = 0
q = deque([src])
visited = {src}
while q:
wavefront_len = len(q)
for _ in range(wavefront_len):
u = q.popleft()
for v in adj_list[u]:
if v in visited:
continue
dist[v] = dist[u] + 1
visited.add(v)
q.append(v)
return dist
Shortest path distances from source with non-negative edge weight
We can extend BFS to accommodate non-negative edge weight (or length) in the following way:
Step 1
Replace an edge having weight
with
copies of 1-weight edges by inserting
virtual nodes. All edges of this new graph has edge length = 1, so we can use BFS.
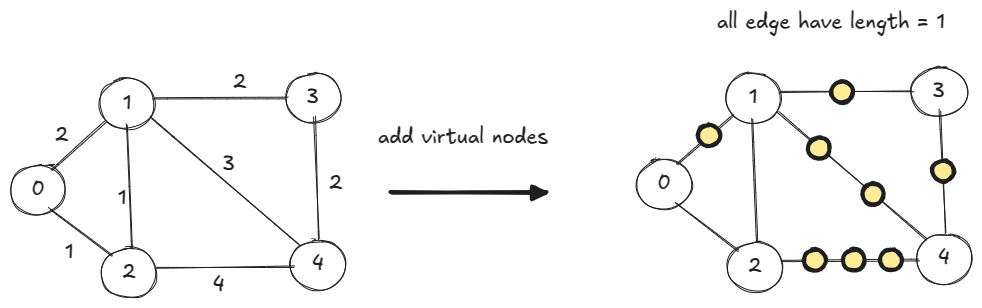
It can be very inefficient. For the below graph, BFS will spend about 100 iterations processing virtual nodes before it finds . Then it spends another 50 iterations (via original edge
) to find
.

Step 2
We just need to reach in one iteration.
Note that, if we ran BFS on the original graph, without inserting virtual nodes, would have been the nearest (from the source) node in the queue. So, while the plain BFS always processes the front of the queue, here we need to process the node in the queue that is nearest to source. We replace the plain queue with a min-priority queue with distance from source as key. This is Dijkstra’s shortest path algorithm.
Our min-priority queue should support efficient key update, because, like in the above example, we might find shorter paths through other nodes.
Python‘s heapq does not support efficient update of key. So, we use UpdatableMinHeap, which wraps a balanced binary search tree (SortedSet) and a dict.
Time: Heap length is . There are
pushes and
pops. For each edge we may get an update, so
updates. Overall time,
.
Space: .
from sortedcontainers import SortedSet
class UpdatableMinHeap:
def __init__(self):
self.sset = SortedSet()
self.map = {}
def push(self, item, priority):
new_entry = (priority, item)
self.sset.add(new_entry)
self.map[item] = new_entry
def pop(self):
_, item = self.sset.pop(0)
del self.map[item]
return item
def update(self, item, priority):
try:
old_entry = self.map[item]
del self.map[item]
self.sset.remove(old_entry)
except KeyError:
raise ValueError(f"No '{item}' to update")
updated_entry = (priority, item)
self.push(item, priority)
def __bool__(self):
return bool(self.sset)
def shortest_path_distance_dijkstra(src, adj_list) -> Dict[str, int]:
dist = {u: float("inf") for u in adj_list}
dist[src] = 0
visited = {src}
q = UpdatableMinHeap()
q.push(src, 0)
while q:
u = q.pop()
for v, w in adj_list[u]:
d_v = dist[u] + w
if v in visited:
if d_v < dist[v]:
dist[v] = d_v
q.update(v, d_v)
continue
visited.add(v)
dist[v] = d_v
q.push(v, d_v)
return dist
adj_list_weighted = {
"A": [("B", 4), ("C", 2)],
"B": [("C", 3), ("D", 2), ("E", 3)],
"C": [("B", 1), ("D", 4), ("E", 5)],
"D": [],
"E": [("D", 1)]
}
dist = shortest_path_distance_dijkstra("A", adj_list_weighted)
print(f"{dist=}")
# dist={'A': 0, 'B': 3, 'C': 2, 'D': 5, 'E': 6}
Dijkstra’s shortest path is almost the same as the plain BFS shortest path, except three differences:
- Weight: Not always 1.
- Queue: Priority queue vs. plain queue
- Update: Even if a vertex has been visited, in Dijkstra, if we find a better distance, we allow update.
The assumption of non-negative edge weight is used during update. We update without checking if it is in the heap:
if (d_v := dist[u] + w) < dist[v]:
dist[v] = d_v
q.update(v, d_v)
With non-negative edge weights, can be true only if
is closer to the
src than . As a result, these
‘s will be popped before
. Once
is popped, no
exists for which
can be true. As a result, we would not be trying to update a non-existent
.
With the below graph with negative edge weights, the update will fail. Because, has a better update from
which is further away from the
src. So, when we try to update dist["B"] with dist["D"] + (-4), has already been popped and we get
ValueError:

adj_list_weighted_neg = {
"A": [("B", 4), ("C", 2)],
"B": [],
"C": [("B", 1), ("D", 3)],
"D": [("B", -4)]
}
dist = shortest_path_distance_dijkstra("A", adj_list_weighted_neg)
print(f"{dist=}")
# ValueError: No 'B' to update.
Shortest path distances from source with negative edge weight
Looking at Dijkstra’s update, the loop maintains the invariant: dist[v] is either overestimate or exactly correct — it cannot go too low. It cannot go too low because, updates are happening along some neighbor aka along some valid path.
if (d_v := dist[u] + w) < dist[v]:
dist[v] = d_v
q.update(v, d_v)
So, updates are actually safe. What if we just relaxed: if does not exist in the queue, it’s ok, just update
dist[v] anyway.
The relaxed Dijkstra works for the below graph with negative weight:

def shortest_path_distance_dijkstra_relaxed(src, adj_list) -> Dict[str, int]:
dist = {u: float("inf") for u in adj_list}
dist[src] = 0
q = UpdatableMinHeap()
q.push(src, 0)
visited = {src}
while q:
u = q.pop()
for v, w in adj_list[u]:
d_v = dist[u] + w
if v in visited:
if d_v < dist[v]:
try:
q.update(v, d_v)
except ValueError:
pass
finally:
dist[v] = d_v
continue
dist[v] = d_v
visited.add(v)
q.push(v, d_v)
return dist
adj_list_weighted_neg = {
"A": [("B", 4), ("C", 2)],
"B": [],
"C": [("B", 1), ("D", 3)],
"D": [("B", -4)]
}
dist = shortest_path_distance_dijkstra_relaxed("A", adj_list_weighted_neg)
print(f"{dist=}")
# dist={'A': 0, 'B': 1, 'C': 2, 'D': 5}
However, even the relaxed Dijkstra cannot correctly find the shortest distances for the below graph with negative edge weights:
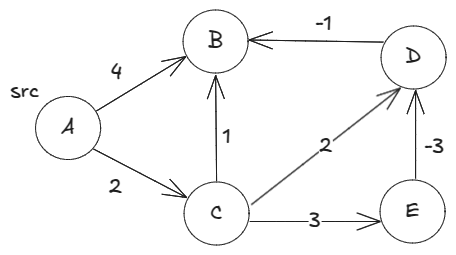
has a shortest path distance of 1 via
and
, but the relaxed Dijkstra gives
dist["B"] = 3.
adj_list_weighted_neg2 = {
"A": [("B", 4), ("C", 2)],
"B": [],
"C": [("D", 2), ("E", 3)],
"D": [("B", -1)],
"E": [("D", -3)]
}
dist = shortest_path_distance_dijkstra_relaxed("A", adj_list_weighted_neg2)
print(f"{dist=}")
# dist={'A': 0, 'B': 3, 'C': 2, 'D': 2, 'E': 5}
The issue is: since is furthest from the
src, it is processed in the last iteration. As a result, it barely had time to update dist["D"] along the edge and misses updating
dist["B"] along the edge .
Letting enough updates happen will ensure all distances have settled to their minimum values. The longest shortest path in a vertex graph has
edges. So, we shall let each edge be updated
times. This is Bellman-Ford algorithm.
Time: .
def shortest_path_distance_bellman_ford(src, adj_list) -> Dict[str, int]:
dist = {u: float("inf") for u in adj_list}
dist[src] = 0
vertex_count = len(adj_list)
for _ in range(vertex_count-1):
for u in adj_list:
for v, w in adj_list[u]:
dist[v] = min(dist[v], dist[u]+w)
return dist
adj_list_weighted_neg2 = {
"A": [("B", 4), ("C", 2)],
"B": [],
"C": [("D", 2), ("E", 3)],
"D": [("B", -1)],
"E": [("D", -3)]
}
dist = shortest_path_distance_bellman_ford("A", adj_list_weighted_neg2)
print(f"{dist=}")
# dist={'A': 0, 'B': 1, 'C': 2, 'D': 2, 'E': 5}
Note, the shortest path distance question is ill-posed for a graph with a negative cycle — a cycle whose edge-weights sum up to a negative number. Because, we can always find a shorter path by circling along the negative cycle. If we let all edges be updated time instead of Bellman-Ford’s
times, then in the last iteration an update will still occur if and only if the graph has a negative cycle. We can use this fact to piggy-back on the distance finding to detect the presence of a negative cycle.
Shortest path distance for Directed Acyclic Graph (DAG)
A DAG does not have a cycle, so it cannot have negative cycles. Shortest path distance question is never ill-posed for DAGs. Additionally, if we process the edges in the topological order (say left-to-right) of vertices, the edges will always flow rightwards. As a result, we can perform a single update (instead of ) of the edges in topological order and be done.

Time: .
def sorted_topologically(adj_list) -> List[str]:
ind = {u: 0 for u in adj_list}
for u in adj_list:
for v, _ in adj_list[u]:
ind[v] += 1
sources = [u for u, d in ind.items() if d == 0]
topo = []
while sources:
topo.extend(sources)
new_sources = []
for u in sources:
for v, _ in adj_list[u]:
ind[v] -= 1
if ind[v] == 0:
new_sources.append(v)
sources = new_sources
return topo
def shortest_path_distance_dag(src, adj_list) -> Dict[str, int]:
dist = {u: float("inf") for u in adj_list}
dist[src] = 0
for u in sorted_topologically(adj_list):
for v, w in adj_list[u]:
dist[v] = min(dist[v], dist[u]+w)
return dist
In fact, the below graph where relaxed Dijkstra failed and we used Bellman-Ford, is actually a DAG. So, instead of Bellman-Ford’s quadratic time, we could get away with linear time.

adj_list_weighted_neg2 = {
"A": [("B", 4), ("C", 2)],
"B": [],
"C": [("D", 2), ("E", 3)],
"D": [("B", -1)],
"E": [("D", -3)]not reachable from src
}
dist = shortest_path_distance_dag("A", adj_list_weighted_neg2)
print(f"{dist=}")
# dist={'A': 0, 'B': 1, 'C': 2, 'D': 2, 'E': 5}
Leave a comment